Scientific Posters
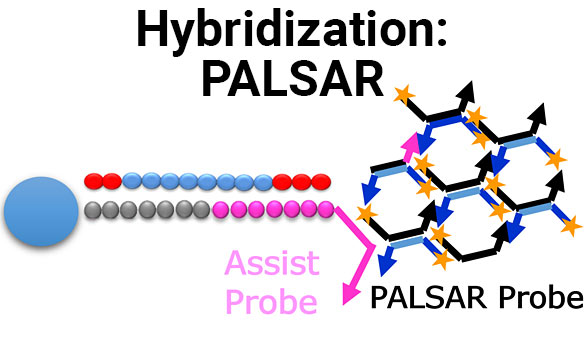
Establishment of PALSAR method for quantification of human cells in mouse kidney
Full Title Establishment of PALSAR method for quantification of human cells in mouse kidney Authors Tetsuo Sekino, Akane Omori and AkiraIdeno Analytical Technology Center for...
Highly selective and sensitive hybridization-based assay for quantification of ASOs drug using signal amplification method
Full Title Highly selective and sensitive hybridization-based assay for quantification of ASOs drug using signal amplification method Authors Yuya Ono1, Tetsuo Sekino1, Takuro Akiya1, Funa...
A Novel ADA Screening Assay for Immunogenicity Testing of Oligonucleotide Drug Using PALSAR Technology
Full Title A Novel ADA Screening Assay for Immunogenicity Testing of Oligonucleotide Drug Using PALSAR® Technology Authors Masako Osawa, Funa Ogawa, Koichi Shibusawa and Akira...
Investigation of the PPB method for oligonucleotide drugs by using the PALSAR
Full Title Investigation of the PPB method for oligonucleotide drugs by using the PALSAR® Authors Rieko Sakai1, Takashi Yamamoto1, Masako Osawa2, Funa Ogawa2, Shun Kumagaya3,...
Establishment of the PPB method for antisense oligonucleotides (ASOs) by using the PALSAR method
Full Title Establishment of the PPB method for antisense oligonucleotides (ASOs) by using the PALSAR® method Authors Rieko Sakai1, Takashi Yamamoto1, Masako Osawa2, Funa Ogawa2,...
Effects of Monocyte Chemoattractant Protein-1, Macrophage Inflammatory Protein-1α and Interferon-α2a on P450 Enzymes in Human Hepatocytes in Vitro
Some immunomodulatory drugs, such as tilsotolimod, stimulate the innate immune system to release cytokines that may change expression of drug-metabolizing enzymes. In drug development, regulation of multiple pro- and anti-inflammatory cytokines by Toll-like receptors (TLR) has gained attention in parallel to targeting the therapeutic potential of these receptors. The aim of this study was to establish whether MCP-1, MIP-1α or IFN-α2a were responsible for the effects of tilsotolimod-stimulated plasma on CYP1A2 and CYP2B6 mRNA and enzyme activity levels...
Toxicity evaluation by high content analysis of 3D cultured primary human hepatocytes
Full Title Toxicity evaluation by high content analysis of 3D cultured primary human hepatocytes Authors Toshimasa Ito, Kazuhiro Fujimoto, Ryo Fujino, Masato Kobayashi, Miki Fujishima,...
Comparison Between the Final US FDA, Japan PMDA and EMA In Vitro DDI Guidance
Comparison Between the Final US FDA (2020), the Final Japan PMDA (2018), and Final EMA (2013) In Vitro DDI Guidance Documents: Are We Finally Harmonized?...
The evaluation of induction and inhibition potency of various drugs on CYP3A4 and OATP1B1/1B3
Full Title The evaluation of induction and inhibition potency of various drugs on CYP3A4 and OATP1B1/1B3 Authors Ryota Takeuchi, Rena Kusano, Tomoko Sasai, Takami Sarashina,...
CYP1A2 enzyme activity and protein abundance in normal and diseased pediatric livers
Full Title CYP1A2 enzyme activity and protein abundance in normal and diseased pediatric livers Authors M. Czerwinski, B. Ewy, A. Kats, M. T. Pritchard, S....